In the Moncla lab, we are interested in how viruses emerge in human populations, and transmit between them. We draw on tools from phylodynamics, virology, and population genetics to understand how viruses evolve within individuals, between populations, and across continents. The ultimate goal of our work is to better understand viral evolution and transmission so that we can prevent new outbreaks from occurring and mitigate the toll of endemic viral transmission. Although our lab primarily uses computational methods, we also generate new genomic data and draw on tools from basic virology to validate our computational findings.
Viral emergence and cross-species transmission
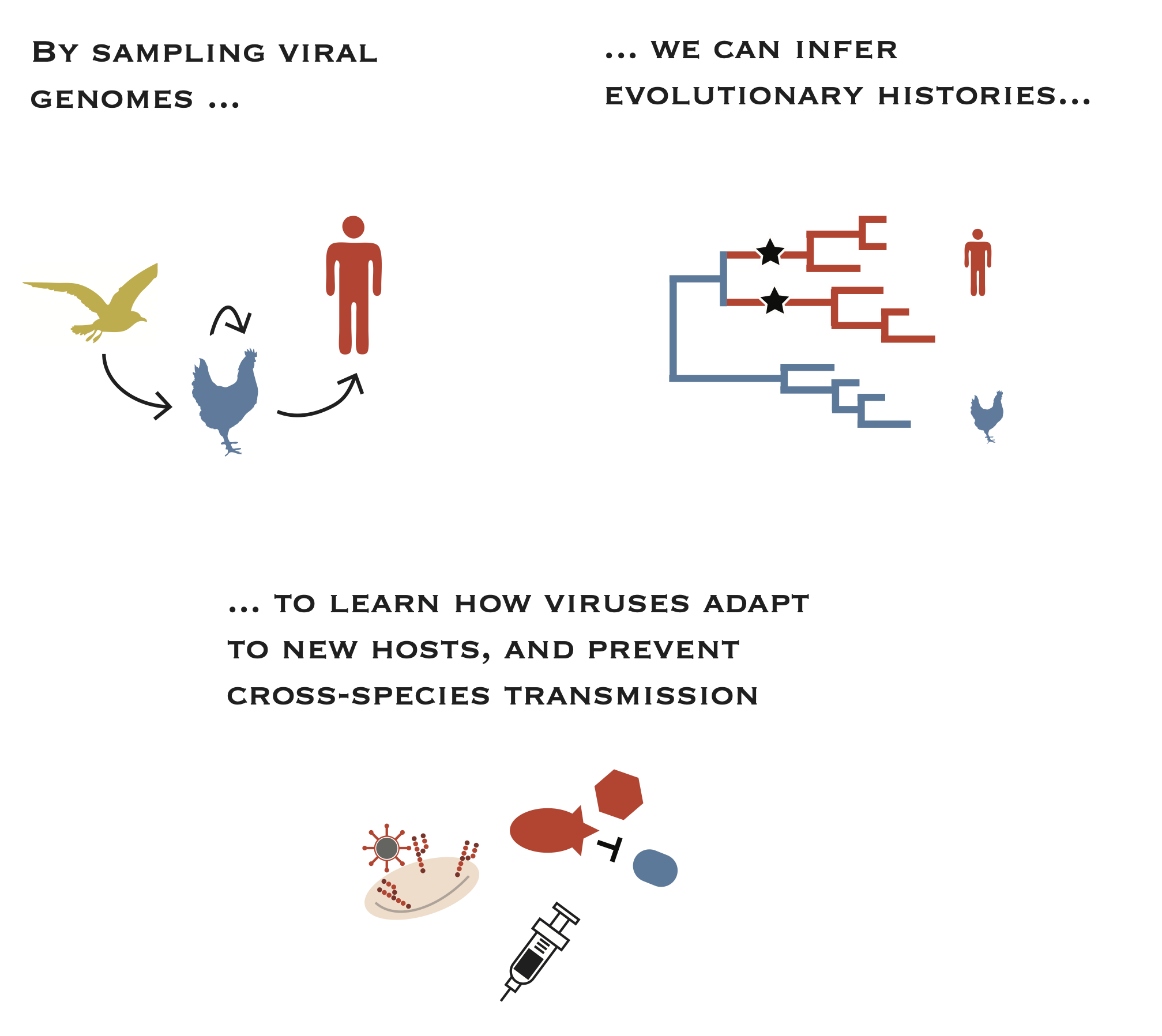
Viruses are constantly crossing species barriers and causing outbreaks in humans. Many new viral pathogens are zoonotic, meaning that they come from animal reservoirs. Most animal viruses are poorly adapted to humans, and must evolve the ability to replicate and transmit among humans. Simultaneously, environmental and epidemiologic factors like habitat encroachment, husbandry practices, and climate can facilitate opportunities for cross-species tranmission by bringing distinct host species together in new ways. We are interested in defining the genetic, epidemiologic, and ecological factors that contribute to cross-species transmission of novel, zoonotic viruses. We are particularly interested in avian influenza viruses, which have repeatedly crossed species barriers and infected humans. By defining the evolutionary pathways by which these viruses adapt to new hosts, we can improve surveillance and vaccine design and improve human health.
Within-host diversity and adaptation
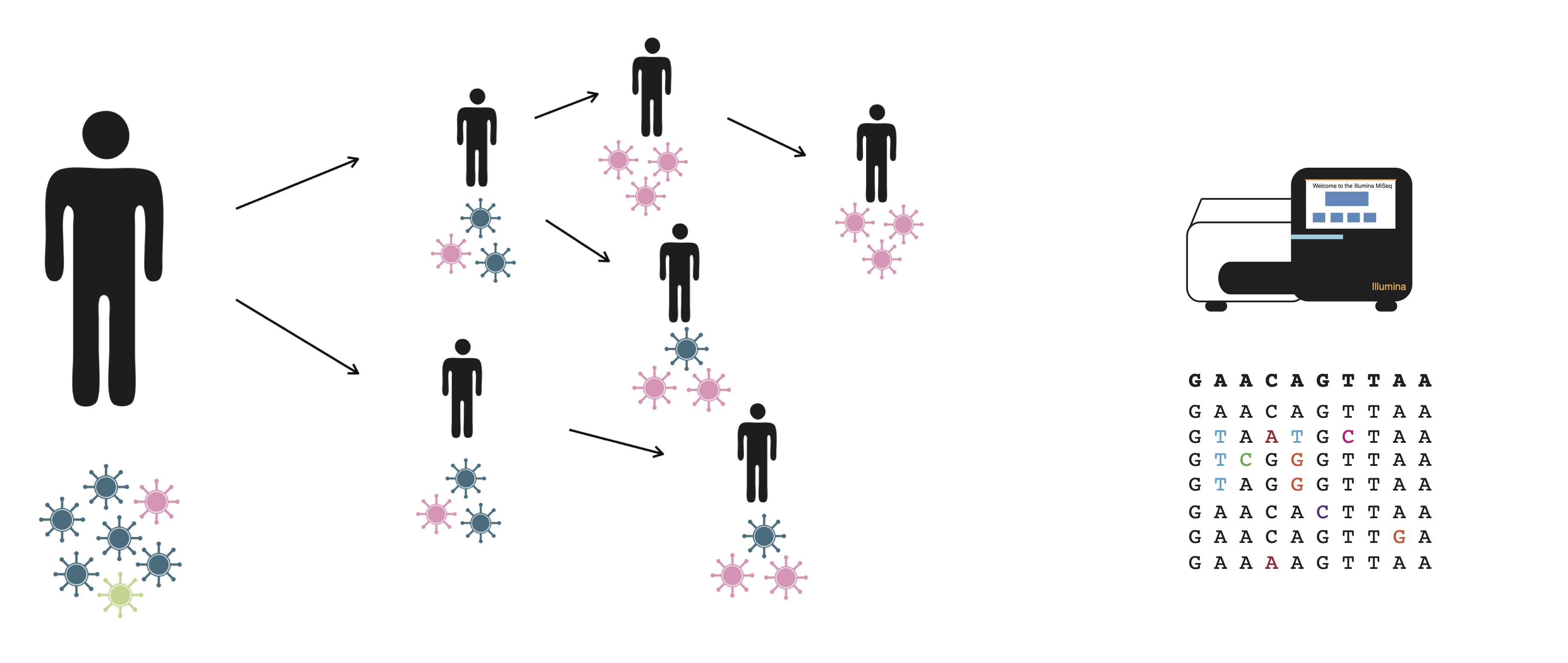
RNA viruses rapidly generate diversity because of their high mutation rates and quick generation times. Because of this, infections with RNA viruses are comprised of a population of viruses that harbors genetic diversity. The diversity that is generated within individuals is then modulated by the process of transmission, during which “transmission bottlenecks” can drastically alter the diversity and makeup of viral populations. By characterizing how viruses acquire mutations during infection, and how the fate of those mutations are shaped by transmission, we can better understand how novel variants arise and spread.
Phylodynamics of endemic human respiratory viruses
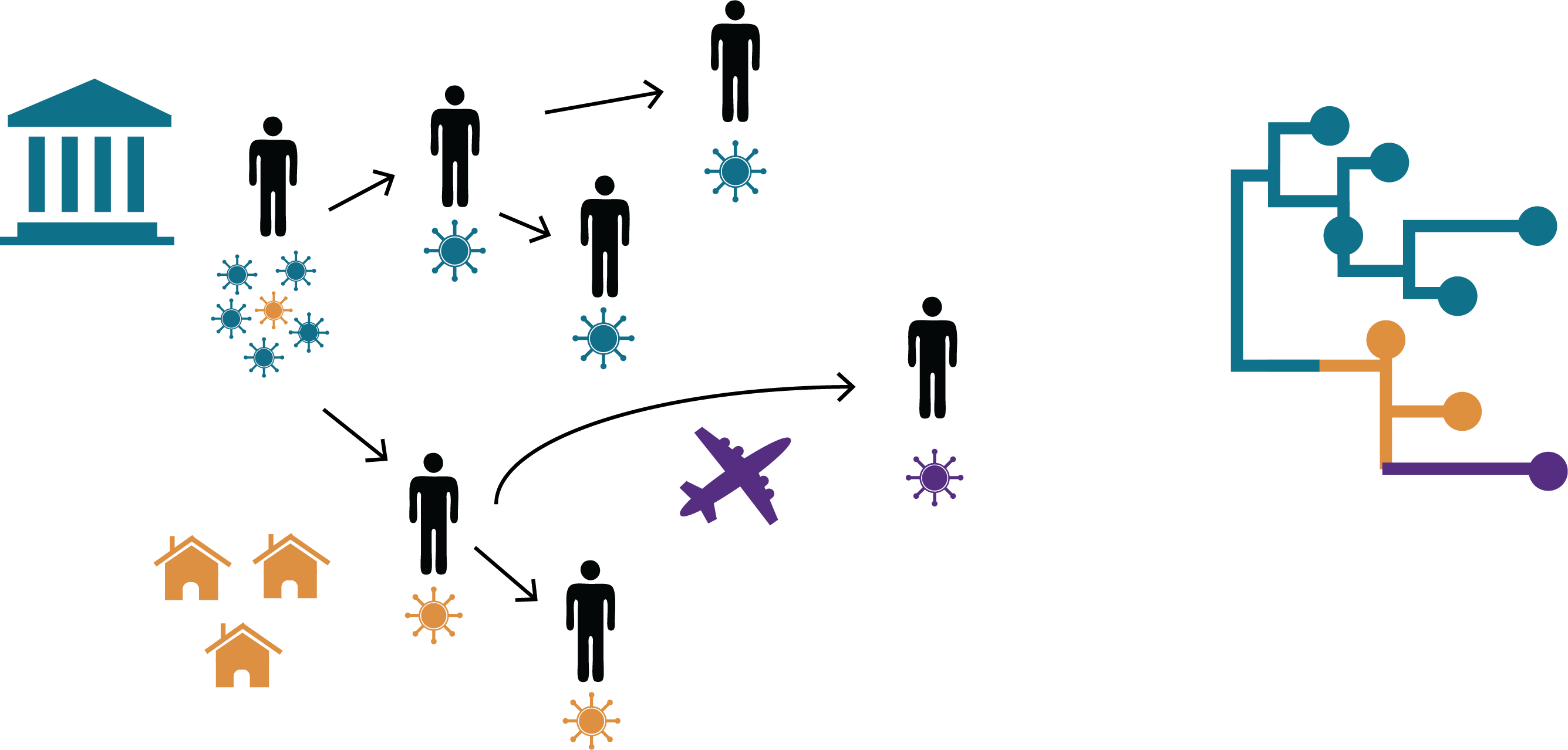
In recent years, viruses that have been previously well-controlled by vaccination have made a resurgence. For endemic human viruses, transmission intensity is impacted by the interplay between population immunity, vaccination campaigns, and human travel, movement, and interaction patterns. We are interested in how endemic RNA viruses move across time, space, and population groups, and how viral transmission is impacted by factors like geography, host immunity, and human contact networks.
Code and data availability
For code and ongoing work, check out our lab github page. We will be keeping scripts and protocols documented there. For Moncla lab custom Nextstrain builds, check out our Moncla lab Nextstrain Group page. We’re tracking a few viruses beyond what is shown on the main Nextstrain site, and those builds are made public here. If you have any questions, don’t hesitate to reach out!