Data and resources
We are intersted in work that lies at the intersection of basic science and public health. We also believe in open science, and are excited by public, interactive data displays. We strive to make our data and code public, and to create tools and visualizations that are useful for academic, public health labs, and interested people from the public.
Nextstrain links:
We are heavily involved in Nextstrain, and built and maintain the avian influenza resources there. Check out global phylogenies for H5Nx, H5N1, H7N9, and H9N2 avian influenza viruses. We update these builds ever month.
If you have avian influenza data that you would like to analyze using Nextstrain resources, check out our tutorial and example data here.
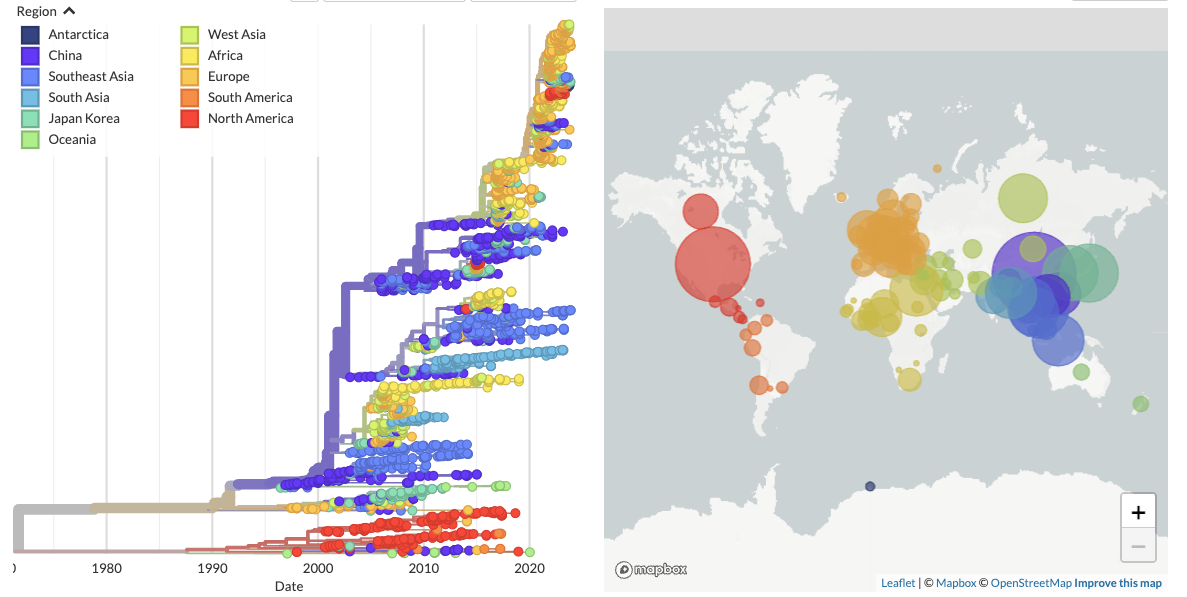
Moncla lab Nextstrain builds
In addition to the resources hosted on the live Nextstrain site, we also maintain custom or work in progress phylogenies for other projects on our Moncla lab Nextstrain Group page. Check out some of our other fun builds there:
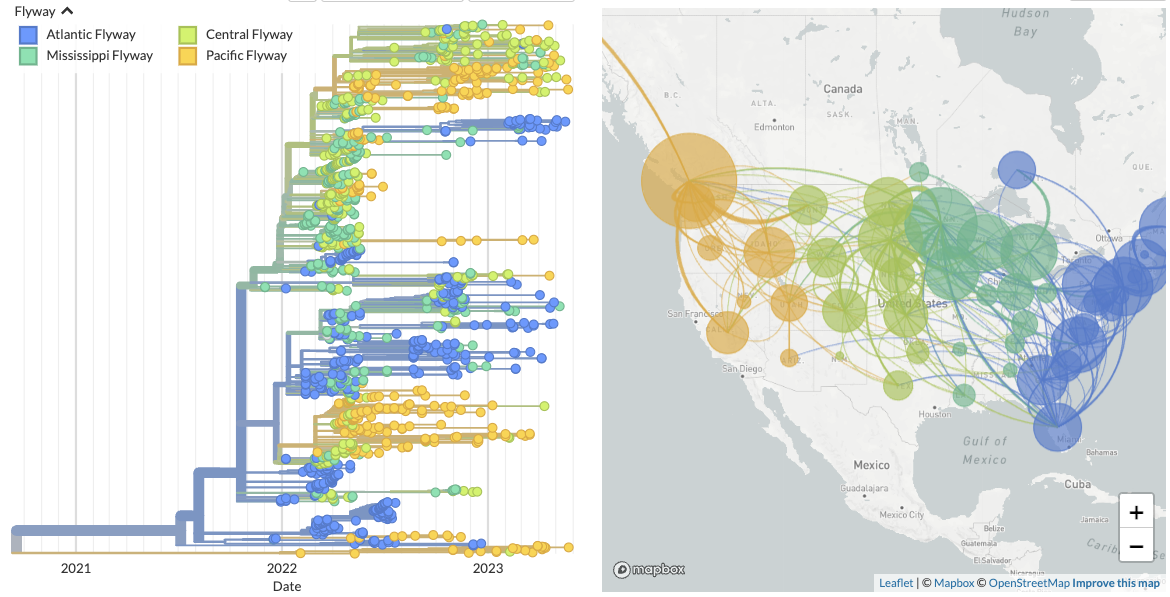
1. Analysis of H5Nx in North America. Since 2022, there has been an enormous outbreak of highly pathogenic H5Nx viruses in the Americas. We built a Nextstrain build that is specifically focused on viruses sampled in the Americas during this outbreak, along with extra features like colors for flyways, avian species, and domestic/wild status.
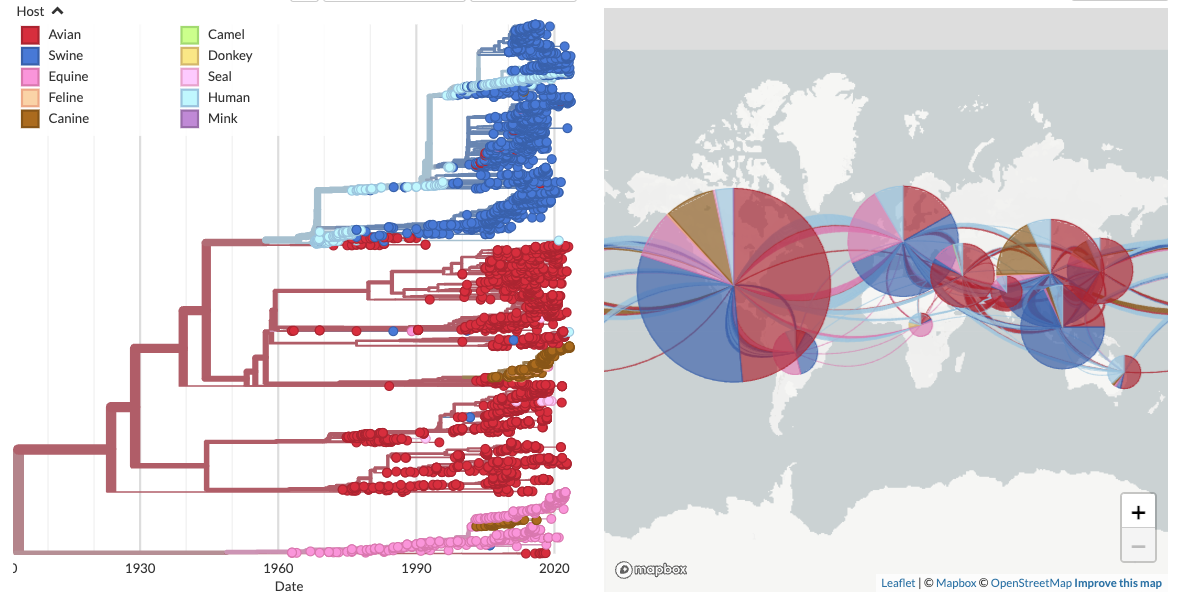
2. Long-term evolution of H3Nx influenza viruses. Avian H3 viruses have jumped into multiple hosts and gone on to circulate endemically, including pigs, horses, humans, and dogs. Maria put together a comprehensive phylogeny of H3Nx evolution across these host species and is using this resource for her PhD work.
Data and code availability:
All of our code for ongoing projects can be found on our lab github page. We will be keeping scripts and protocols documented there. If you have any questions, don’t hesitate to reach out!